Magnaporthe oryzae
Bioactive fungal secondary metabolites and in particular the characterization and regulation of genes essential for their biosynthesis have enormous importance on agrochemistry and pharmacology, e.g. in fungicide development. Fungal secondary metabolites and the regulation of gene clusters responsible for biosynthesis is an actual and timeless research area. Besides the identification of new natural products/secondary metabolites, we aim to identify the genes or gene cluster contributing to their biosynthesis within the genome of fungi. One example of our research area in this field is the identification and characterization of the polyketide synthase encoding gene MoPKS19 of the filamentous phytopathogenic fungus Magnaporthe oryzae, the causing agent of rice blast disease. The gene MoPKS19 was found to be essential for the production of the phytotoxic polyketide pyriculol in M. oryzae. The transcript abundance of MoPKS19 correlates with the biosynthesis rate of pyriculol in a time-dependent manner. Furthermore, gene inactivation of MoPKS19 resulted in a mutant ΔMopks19 unable to produce pyriculol. Neither pyriculol, nor the derivatives thereof were found in axenic cultures of the mutant. Inactivation of a putative oxidase encoding gene MoC19OX1, which was found to be located in the genome close to MoPKS19, resulted in the mutant ΔMoC19ox1 exclusively producing the reduced dihydro-derivatives of pyriculol. In contrast, overexpression of MoC19OX1 resulted in a mutant strain solely producing pyriculol. The MoPKS19 cluster, furthermore, comprises two transcription factors MoC19TRF1 and MoC19TRF2, which were both found individually to act as negative regulators repressing gene expression of MoPKS19.
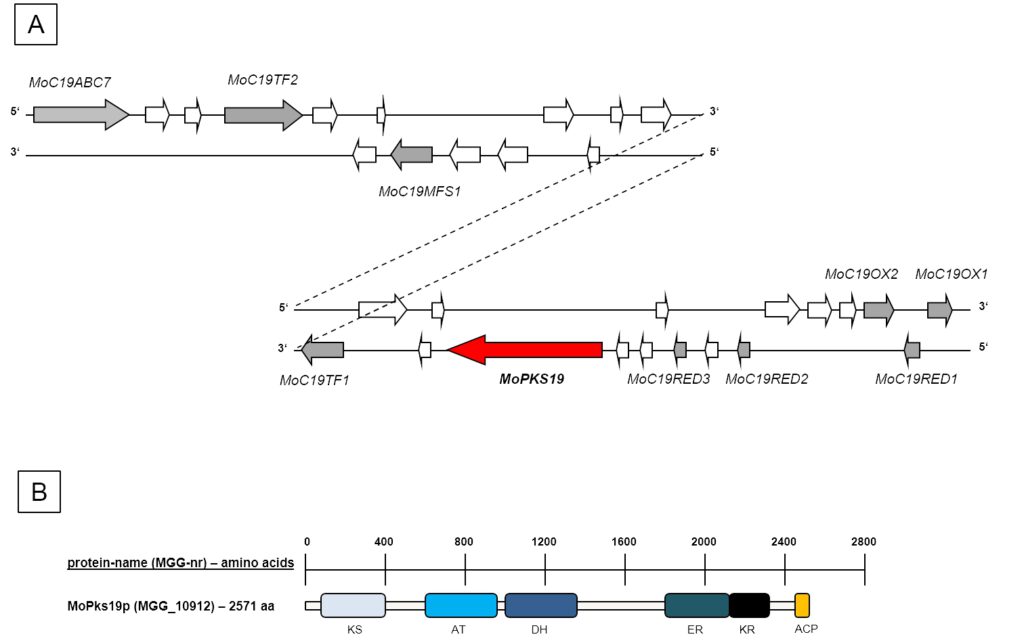

Figure 1: Schematic presentation of the MoPKS19 gene cluster in the genome of MoWT. [A] A scheme of 30 ORFs neighbouring the MoPKS19 gene is presented. The genes are shown with their position either on the “sense (5’-3’)” or the “antisense (3’-5’)” strand. White arrows imply genes with no obvious functions with significant use in this study (e.g. hypothetical proteins without predicted protein domains). [B] Schematic presentation of protein domains of MoPks19p. KS = ß-ketoacylsynthase, AT = acyltransferase, DH = dehydratase, ER = enoylreductase, KR = ketoreductase, ACP = ACP domain.
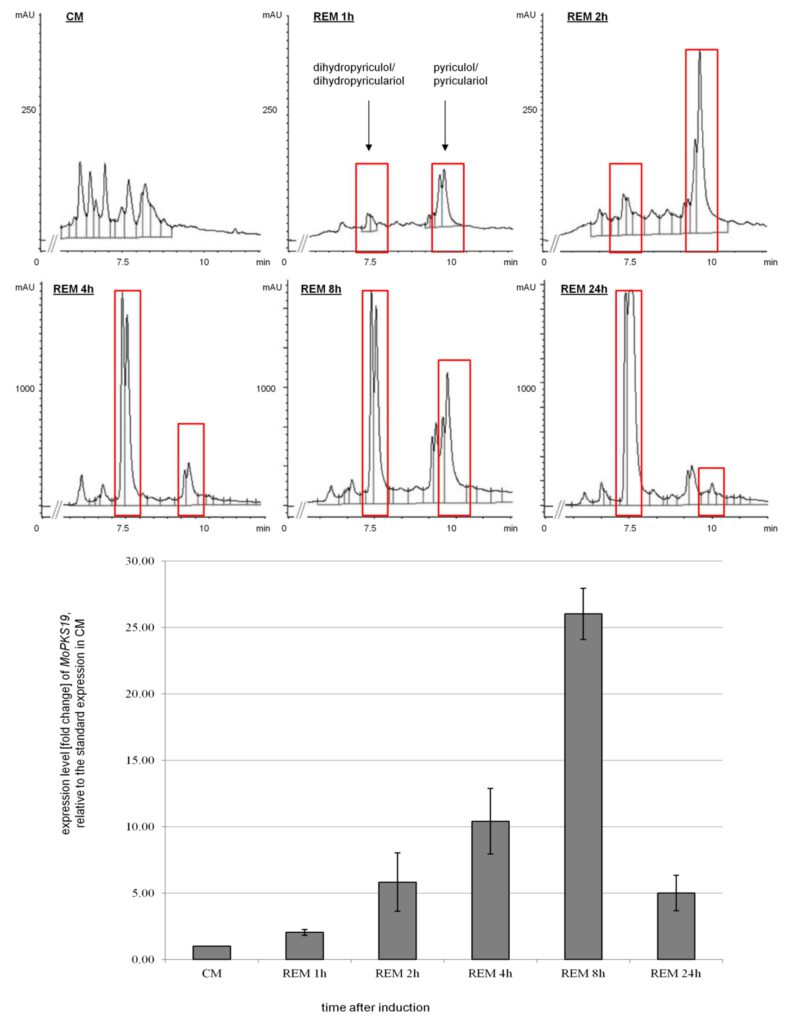

Figure 2: Time-dependent HPLC analysis of culture filtrate extracts from MoWT in correlation to qRT-PCR analysis of the expression level from the MoPKS19 gene. The M. oryzae cultures were grown as described in experimental procedures. Samples were taken 1, 2, 4, 8 and 24 h after transfer to REM. The HPLC analysis was conducted from extracts of the culture broth and RNA was isolated from the mycelium samples. The results of transcript abundance in REM were given relative to quantification in CM. The experiments were conducted in triplicates. CM = complete medium, REM = rice-extract medium, mAU = milli-absorption units.
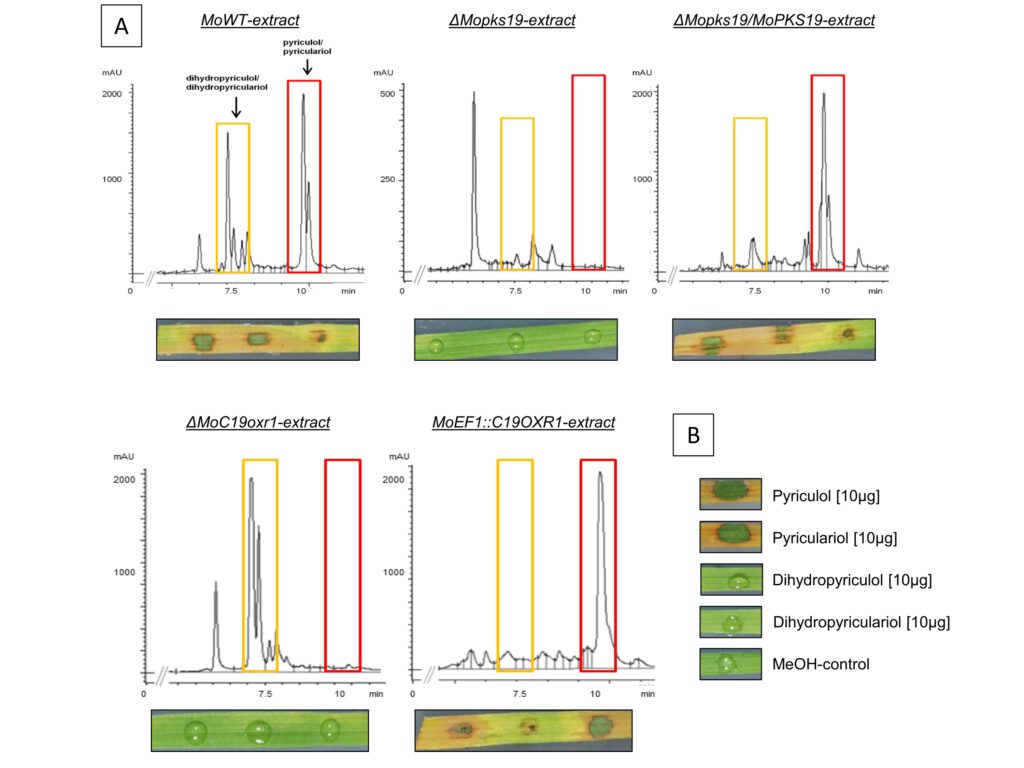

Figure 3: Analysis of PK in culture filtrate extracts from Magnaporthe oryzae and phytotoxic activity of extracts and pure compounds. [A] HPLC chromatograms of extracts from MoWT, ΔMopks19, ΔMopks19/MoPKS19, ΔMoC19OXR1 and the overexpression mutant MoEF1::C19OXR1. The M. oryzae cultures were grown as described in experimental procedures. Samples were taken 8 h after transfer to REM. The HPLC analysis was conducted from extracts of the culture broth (210 nm wavelength). mAU = milli-absorption units. Phytotoxic activity of the extracts towards rice leaves is shown below each chromatogram. The assays were conducted as described in experimental procedures. [B] Phytotoxicity was monitored for the pure compounds under equal conditions.
Related publications
Unravelling the biosynthesis of pyriculol in the rice blast fungus Magnaporthe oryzae.
S. Jacob, T. Grötsch, A. J. Foster, A. Schüffler, P. H. Rieger, L. P. Sandjo, J. C. Liermann, T. Opatz, and E. Thines. Microbiology. 2016, in print. doi:10.1099/mic.0.000396
