Rapid adaptation of signaling networks in the fungal pathogen Magnaporthe oryzae
The question of how the remarkably diverse array of eukaryotic signaling networks has evolved is of great scientific relevance. Evolutionary adaptation of living organisms is commonly thought to be the result of processes that acted over long periods of time. Our research activity in evolutionary processes is motivated by recent observations that microorganisms are able to rapidly adapt to new environments and establish stable phenotypes by natural selection, even within few generations. We found the filamentous rice blast fungus Magnaporthe oryzae to rapidly rewire signal transduction required for osmoregulation in several independent “loss-of-function” (lof)-mutants of the High Osmolarity Glycerol (HOG)-pathway upon exposure to salt stress. Adaptation resulted in stable mutants of the model organism being restored in osmoregulation arising as individuals outgrowing from salt-sensitive lof-mutants (Fig.1).
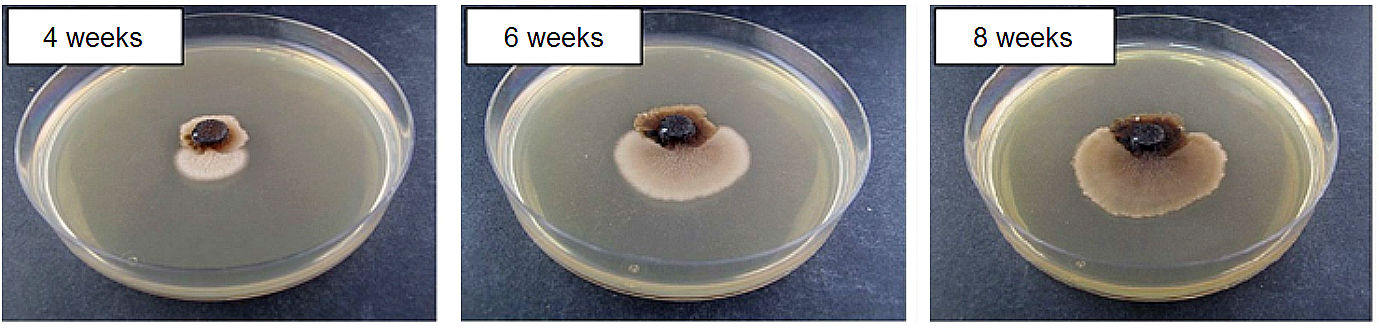 Fig.1: The Magnaporthe oryzae “loss of function” strain ΔMohog1 and the outgrowing adapted strain ΔMohog1(adapted) after cultivation on complete medium including 1 M KCl. The “loss of function” strain ΔMohog1 was grown on complete medium (CM) inclusive 1 M KCl for 4, 6 and 8 weeks at 26 °C. The ΔMohog1 mutant (dark brownish colony) was found to be highly sensitive towards 1 M KCl, whereas the outgrowing ΔMohog1(adapted) strain (bright brownish colony) was able to grow much better.
Fig.1: The Magnaporthe oryzae “loss of function” strain ΔMohog1 and the outgrowing adapted strain ΔMohog1(adapted) after cultivation on complete medium including 1 M KCl. The “loss of function” strain ΔMohog1 was grown on complete medium (CM) inclusive 1 M KCl for 4, 6 and 8 weeks at 26 °C. The ΔMohog1 mutant (dark brownish colony) was found to be highly sensitive towards 1 M KCl, whereas the outgrowing ΔMohog1(adapted) strain (bright brownish colony) was able to grow much better.
Interesting and of high importance is the fact that the observed adaptation phenomenon does not happen with other osmosensitive Magnaporthe mutants which have no relation to the HOG signaling pathway (i.e. ΔMopmk1, ΔMogdp1).
The major compatible solute produced upon salt stress by these rapidly “adapted” strains was found to be glycerol in contrast to arabitol in the wildtype strains (Fig.2).
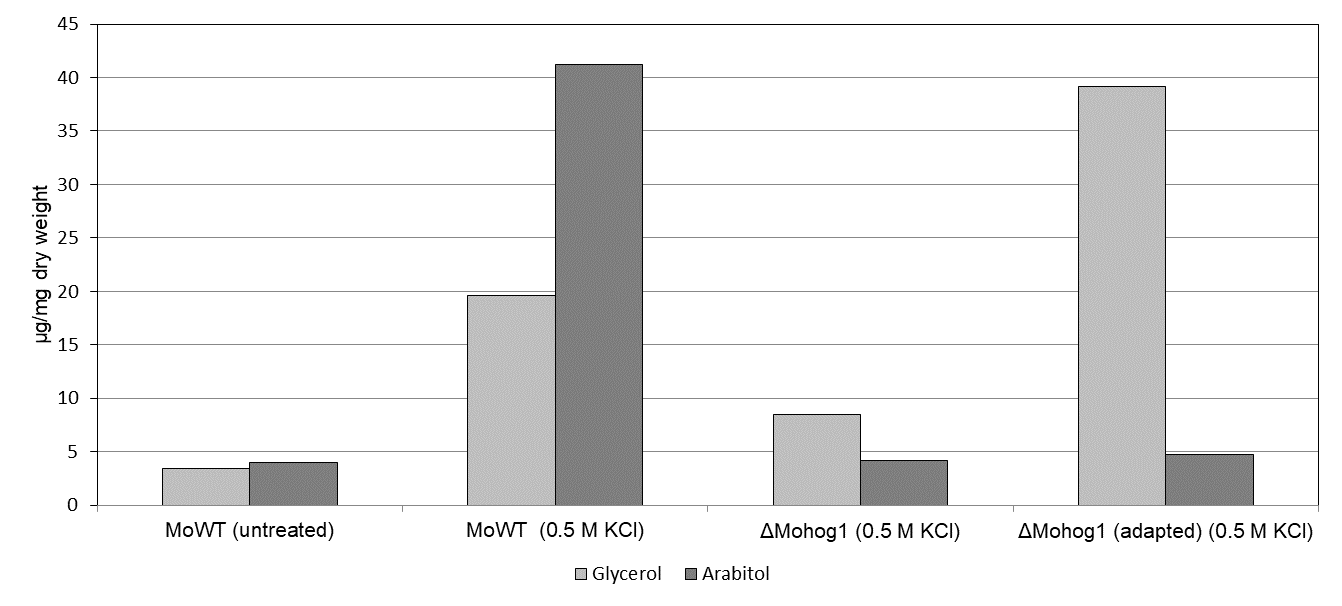 Fig.2: Compatible solute production in the MoWT, the “loss of function”-mutant ΔMohog1 and the “adapted” strain ΔMohog1(adapted). Compatible solute accumulation (glycerol and arabitol) was assayed in mycelium of MoWT, ΔMohog1 and ΔMohog1(adapted) after 7 h salt stress. The mycelium was grown for 72 h in complete medium (CM) before being transferred to CM or CM plus 0.5 M KCl for additional 7 h. Carbohydrates were extracted and quantified by GC/MS and HPLC (HPAEC-PAD).
Fig.2: Compatible solute production in the MoWT, the “loss of function”-mutant ΔMohog1 and the “adapted” strain ΔMohog1(adapted). Compatible solute accumulation (glycerol and arabitol) was assayed in mycelium of MoWT, ΔMohog1 and ΔMohog1(adapted) after 7 h salt stress. The mycelium was grown for 72 h in complete medium (CM) before being transferred to CM or CM plus 0.5 M KCl for additional 7 h. Carbohydrates were extracted and quantified by GC/MS and HPLC (HPAEC-PAD).
These findings lead to the hypothesis that stable adaptation-events under continuously environmental evolutionary pressure enable Magnaporthe oryzae to rapidly restore or modify entire signaling networks. To address this hypothesis, we aim to identify the molecular or biochemical mechanisms of this rapid evolutionary adaption and characterize associated factors and signalling pathways which enable or prevent adaption. We combine expertise of theoretical approaches to integrate sequencing data from genomics and transcriptomics with modern quantitative (phospho)-proteomics techniques (in cooperation with Prof. Stefan Tenzer, Institut für Immunologie, Universitätsmedizin der JGU Mainz, Mainz). In initial experiments over 3000 phosphopeptides were identified and quantified, enabling us to pinpoint key differences in the signaling pathways between MoWT and mutant strains. (Fig. 3).
 Fig.3: Clustering analysis of label-free phosphopeptidomics. Data were obtained from a phosphopeptidomics analysis of MoWT and ΔMohog1 under salt stress conditions. Highly significant differences in the phosphorylation patterns and kinetics were observed. Only the Top70 most regulated proteins are shown.
Fig.3: Clustering analysis of label-free phosphopeptidomics. Data were obtained from a phosphopeptidomics analysis of MoWT and ΔMohog1 under salt stress conditions. Highly significant differences in the phosphorylation patterns and kinetics were observed. Only the Top70 most regulated proteins are shown.
In order to obtain further information regarding the mechanisms of adaptive evolution at genome level, we have already sequenced the gDNA of ΔMohog1 and ΔMohog1(adapted). Based on the sequencing results, we carried out variant calling analysis aiming to compare different events of structural sequence variants (deletion, insertion, substitution). In addition, these data provide the basis to generate hypotheses on putative effects at the protein level. The genomic data set was complemented by RNA-Seq data from sequencing ΔMohog1 and ΔMohog1(adapted) – these NGS results were integrated into the processed gDNA data set. In the focused search for “changes” that could be relevant for rapid evolutionary adaptation, we identified a first set of candidate genes by means of comparative variant calling analysis (GATK, SnpEFF), which are visualized as a VENN diagram (Fig. 4).
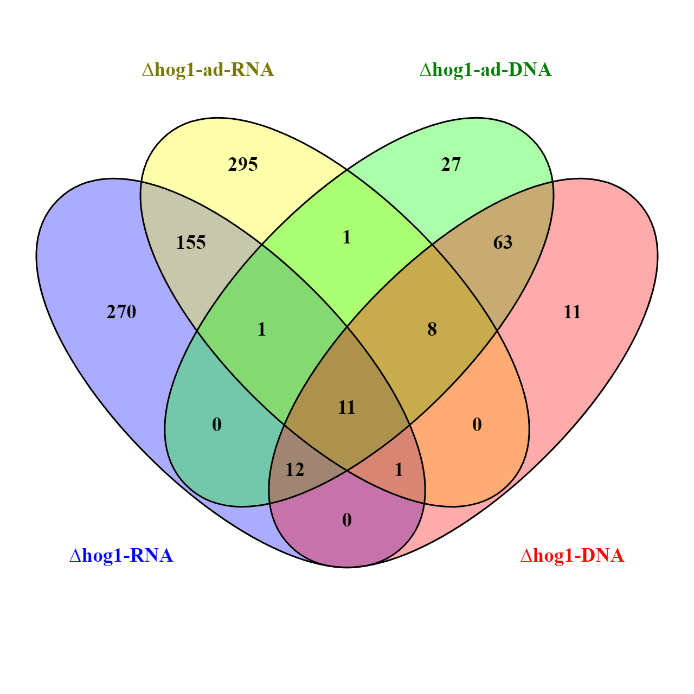 Fig.4: Example of VENN chart of RNAseq- and gDNAseq data of ΔMohog1 and ΔMohog1(adapted). The VENN-diagram is presented to visualize the number of putative candidate genes potentially responsible for rapid evolutionary adaption. A variant calling analysis was conducted based on combinatorial approach employing GATK and SnpEFF.
Fig.4: Example of VENN chart of RNAseq- and gDNAseq data of ΔMohog1 and ΔMohog1(adapted). The VENN-diagram is presented to visualize the number of putative candidate genes potentially responsible for rapid evolutionary adaption. A variant calling analysis was conducted based on combinatorial approach employing GATK and SnpEFF.
Furthermore, we use reversed molecular genetics to validate the candidate genes or even other factors (e.g. phosphorylation patterns) found to putatively promote or constrain rapid evolutionary adaptation.
To our delight, these preliminary work enabled us to join the DFG priority programme SPP1819 “Rapid Evolutionary Adaptation: Potential and Constraints”.
Related publications
Rapid adaptation of signaling networks in the fungal pathogen Magnaporthe oryzae.
S. Bohnert, L. Antelo, C. Grünewald, A. Yemelin, K. Andresen, and S. Jacob.
BMC Genomics 20, 763. doi: 10.1186/s12864-019-6113-3.
