Many of the interesting fungal enzymes are produced by organisms that grow very slowly (e.g. basidiomycetes) or that grow conditions are too expensive for industrial proposes. Therefore, it is often necessary to produce those proteins heterologous in a more suitable organism. Unfortunately, the fungal proteins often cannot be expressed in model organisms like E. coli or yeast since these host organisms are not able to fulfill essential post-translational modifications (e.g. glycosylation, folding, or activation). Furthermore, expression systems in filamentous fungi often have several advantages over bacterial systems for protein production: Besides post-translational modifications, a high-level secretion of enzymes is a common trait of their lifestyle. That feature may in some cases simplify the isolation and thus the purification process of heterologous expressed enzymes.
At the IBWF we have developed several heterologous expression systems in filamentous fungi. Our experience comprises the expression of recombinant proteins under the control of inducible or constitutive promoters, fused to fluorescent markers, signal sequences for secretion, tags for purification or increased solubility.
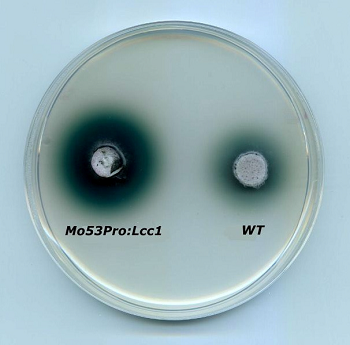
The filamentous fungus Magnaporthe oryzeae used for the heterologous expression of a laccase from Coprinopsis cinerea, compared with the wild tipe. Green color indicates laccase activity.

Purification of a GFP-tagged protein
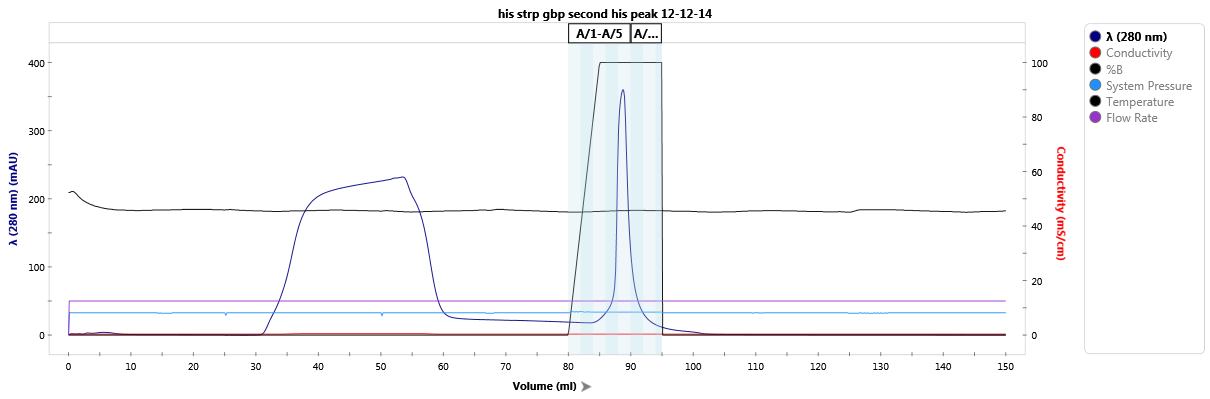
Affinity purification of a StrepTag-fused protein.
Related publications
Magnaporthe oryzae as expression host for the production of the unspecific peroxygenase AaeUPO from the basidiomycete Agrocybe aegerita.
Jacob, S., Bormann, S. Becker, M., Antelo, L., Holtmann, D., Thines, E., 2021
MicrobiologyOpen. doi: 10.1002/mbo3.1229.
