Despite the economic importance of many fungi, the molecular basis regarding the pathogenic differentiation, in planta growth and development has not been investigated in large detail. The remaining paucity regarding the molecular-genetic basis of pathogenicity-related determinants makes particularly the use of next generation sequencing (NGS) technologies, proteomics and metabolomics required to gain insight into different stages of the infection cycle, in order to determine novel directions and strategies for the disease control. Taking advantage of the techniques and tools, one of the main goals of our research is focused on the investigation of molecular mechanisms governing the fungal pathogenesis with emphasis on characterization of critical molecular and cellular processes related to virulence by employing bioinformatic approaches complemented by wet lab analysis. Much of our work is focused on investigation of dynamic biological processes by employing transcriptome analysis using RNA-Seq to get insight into the regulatory processes governing plant-pathogen interaction during the infection process. In addition by employing of structural genomic analysis we attempt to complement and integrate the data obtained from biochemical, histological and “omics” technologies. Furthermore we are interested in development of new strategies for data analysis and data mining aiming to facilitate and improve the biological knowledge discovery.
Recent projects in bioinformatics:
- Mode of action studies based on microarray technology
- De novo genome assembly in fungi
- Fungal genome annotation
- Quantitative RNA-Seq analysis
- Comparative genomics
- Gene enrichment, clustering and data visualization
- Gene regulatory networks
- Metabolic pathways and regulatory signaling systems
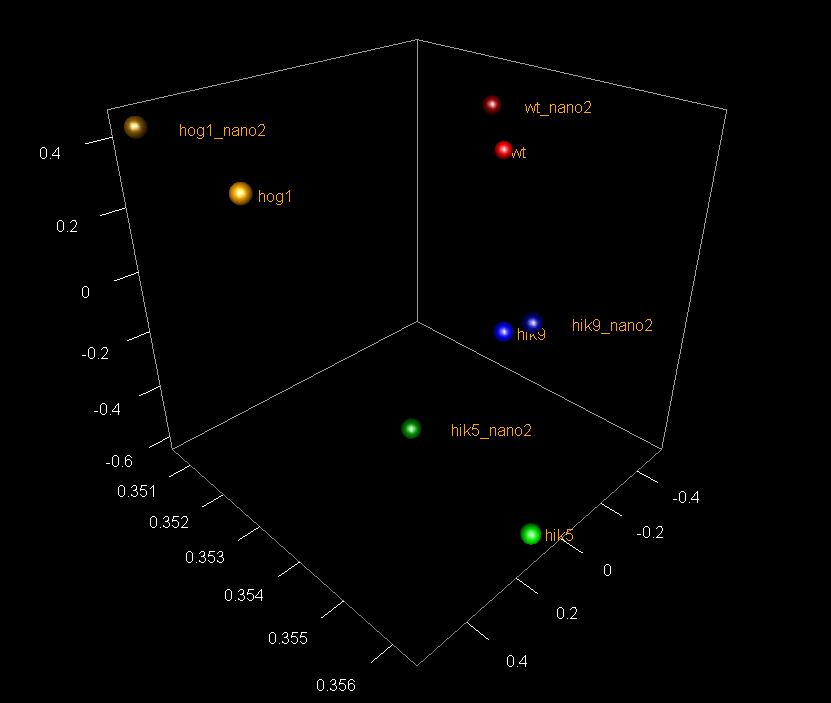
A: Quality control of RNA-Seq data by PCA (click on image for interactive 3D model)
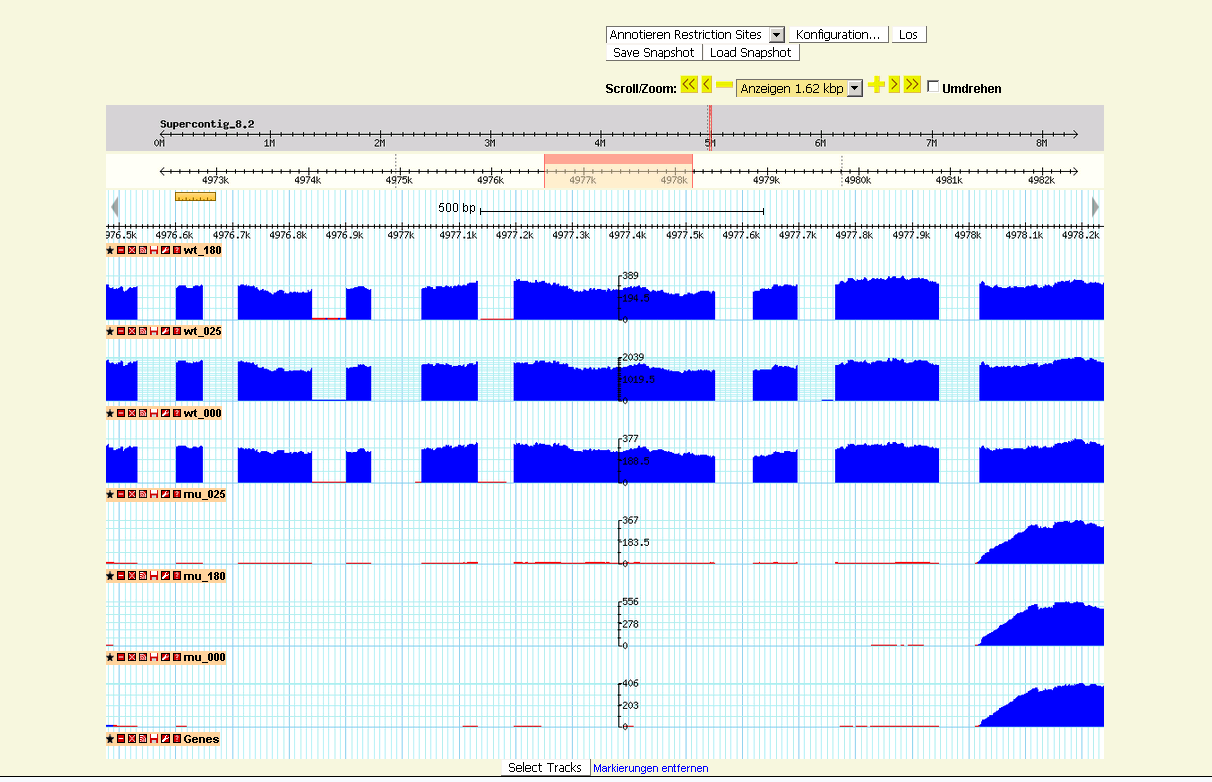
B: RNA-Seq data visualization with gbrowse
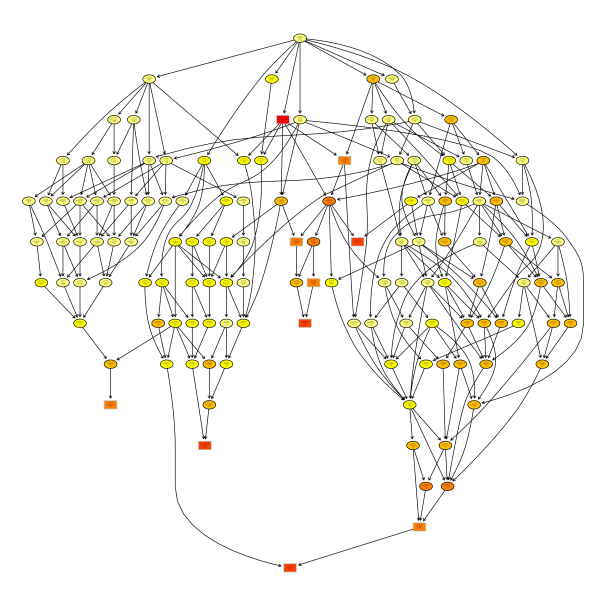
Gene enrichment by topGO clustering
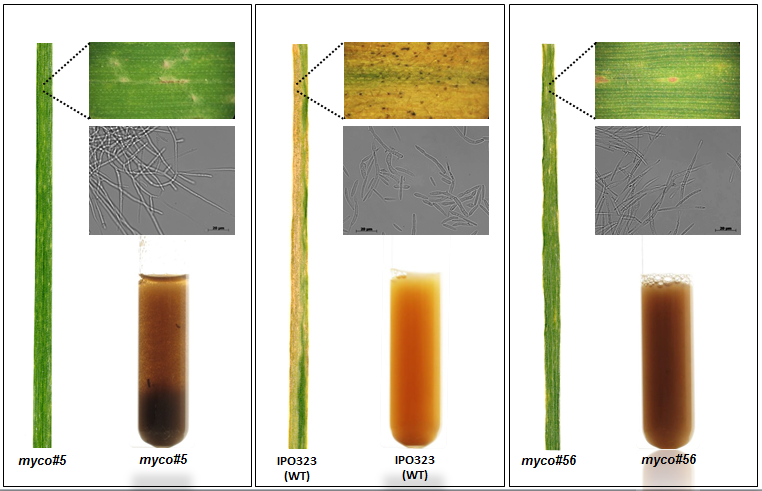
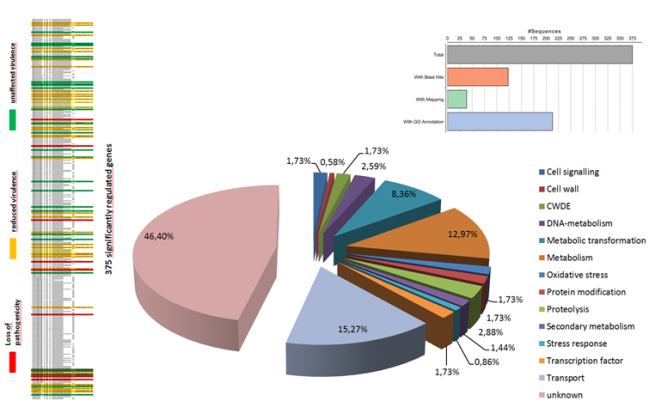
Another research area of our lab is focused on understanding of fungal secondary metabolism and identifying of novel metabolites. Fungi represent a vast source of various natural products which have diverse activities, ranging from antibiotics to lethal poisons. Many of them are synthesized using non-ribosomal peptide synthetase or polyketide synthase pathways. Generally, the natural products are among the most important sources of lead molecules for drug discovery.
With the development of NGS technologies and other “omics tools”, the field of natural products research is currently undergoing a shift in paradigms. While, for decades, mainly analytical and chemical methods gave access to this group of compounds, nowadays genomics-based methods offer complementary approaches to identify and characterize such molecules. Nowadays, “omics” approaches offer complementary access to natural products; by identifying natural product/secondary metabolite biosynthetic gene clusters, it is possible to assess the genetic potential of producer strains and to more effectively identify previously unknown metabolites.
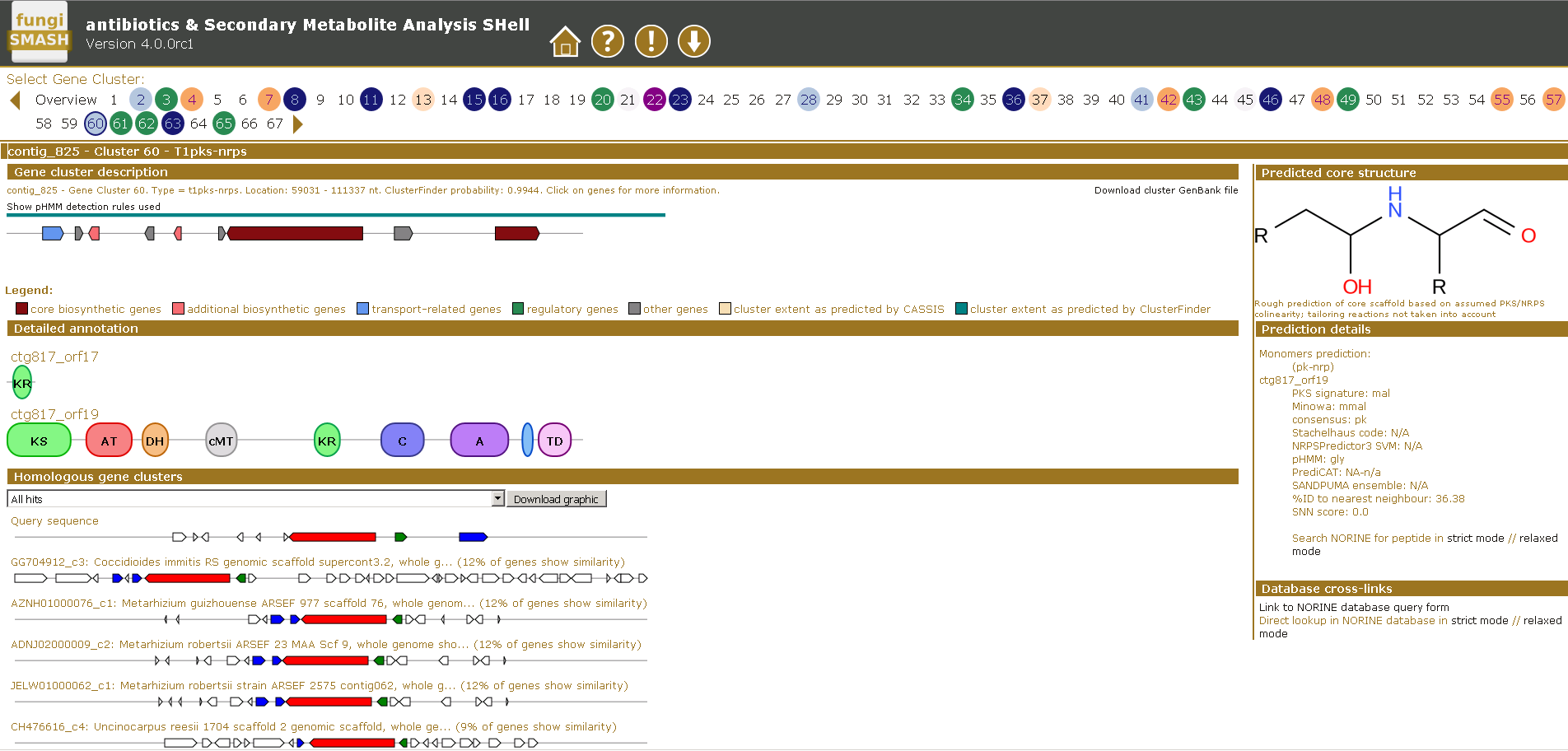
Many fungi contain over 40 of these clusters, with each having the capacity to make multiple novel compounds. However the biological role of these compounds, or indeed their identity, for the most part remain unknown. By exploiting different bioinformatics approaches we are focused on elucidation of biosynthetic gene clusters to identify the novel metabolites and discover their roles in causing disease.
